One of the arguments I've heard from creationists during debates and presentations over the past few years is that analyses of protein sequences do not support the scientific view that one-celled organisms, invertebrates, fish, and tetrapods slowly evolved from each other in this sequencethe less complex forms increasing in complexity slowly over the eons. The source of this stunning pronouncement (at least, I was stunned when I first heard it) is Michael Denton's book Evolution: A Theory in Crisis (1985). When Denton's protein "data" are presented by a skilled debater, this argument sounds very plausible; in fact, it's not very good science and provides yet another example of the convoluted creationist reasoning we have become all too familiar with. Denton's book is rife with twisted data and half-truths, much of it old hat. Since a great deal of Denton's material has already been addressed in Creation/Evolution and elsewhere, I will restrict my comments to Denton's notions about protein sequences.
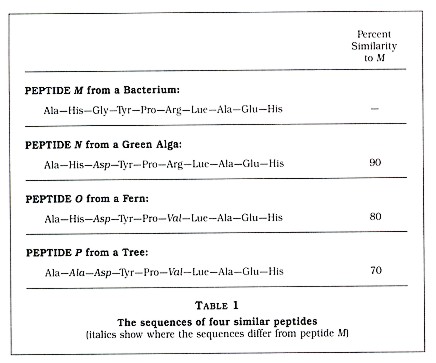
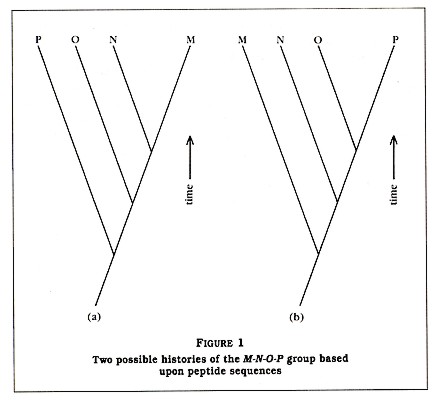
The technique of using protein sequences for re-creating evolutionary relationships is now over twenty years old. In principle, it is assumed that organisms with proteins of similar structure are more closely related than organisms with proteins that are very different. This is best explained by a simple example in which I have substituted small peptides (compounds containing two or more amino acids linked by carboxyl groups) for the larger proteins. Suppose you have sequenced a particular peptide in an organism designated as M. (By sequenced, biologists mean that they have determined the order of amino acids comprised by that peptide.) Then you sequence the same peptide in three other organisms: N, 0, and P (see Table 1). We see that N has the same amino acids as peptide M at 90 percent of the sites. In addition, we see that 0 and P are 80 percent and 70 percent similar to M, respectively. Also note that the new amino acid that is seen in N is also seen in 0 and that both of the new amino acids seen in 0 are also seen in P. Based upon these results, one could reconstruct two possible histories for the four species, as seen in Figure 1.
Which of these two histories is the correct one? Based only upon the amino acid data, it is impossible to say; however, normally other data (paleontological, morphological, developmental, and biochemical) are available. In my example, I have assigned peptide M to a bacterium, peptide N to a green alga, peptide 0 to a fern, and peptide P to a tree (see Table 1). Based upon a good deal of published data, we will assume that the history depicted in Figure 1(a) is more likely to be correct than the history depicted in Figure 1(b). Thus, Figure 1(a) indicates that, at some time in the past, species M and N shared a single common ancestor species but a speciation event resulted in two new species, which evolved slowly over time into the modern M and N species. Since the peptides 0 and P are more dissimilar from peptide M than peptide N is, we can assume that the ancestors of 0 and P branched away earlier from the line leading to M. It is very important to understand that, although P separated from the O-N-M group in the more distant past, it has continued to evolve since that split.
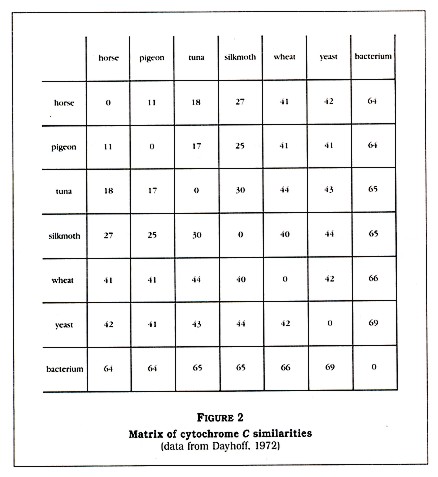
Denton generated a matrix of protein sequences from Dayhoffs Atlas of Protein Sequence and Structure (1972). Again, for the sake of simplicity, I have shortened the matrix a bit (see Figure 2, p. 4). The numbers in this matrix are the percentage of sequence differences; that is, a horse is zero percent different in cytochrome C sequence from itself but about 18 percent different in sequence from a tunafish. Denton calls the reader's attention to the last column, which shows the differences between a bacterium (a prokaryote) and other forms of life (all eukaryotes).
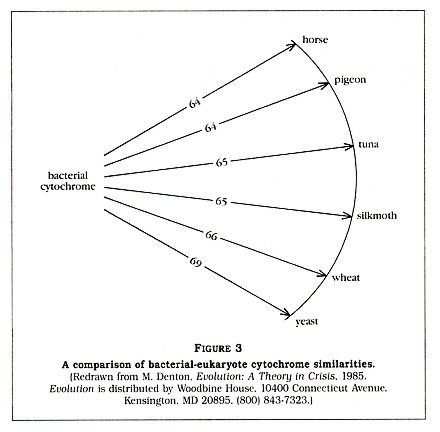
The fact that the eukaryotes may vary greatly amongst themselves (for example, yeast and a silkmoth are 44 percent different) but are all 64 to 69 percent different from the prokaryote is very strange, according to Denton. He tells us that "no eukaryotic cytochrome is intermediate between the bacterial cytochrome and the other eukaryotic cytochromes." Denton has clearly impressed himself with this discovery, calling it "one of the most astonishing findings of modern science." He then generates a diagram (Figure 3, p. 5) to show us that all of the listed eukaryotes are approximately equidistant, in terms of cytochrome C structure, from the bacterium. Amazingyes?
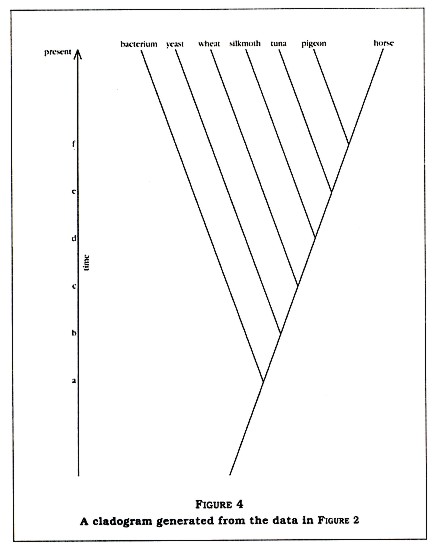
No! Not from the standpoint of evolutionary science. In fact, I would be very surprised to see anything else. Let's draw out a cladogram (sort of a family tree) of the organisms in the matrix based upon conventional evolutionary theory (see Figure 4, p. 6). Notice that at point a on the time line there was a speciation event separating the bacterium from the eukaryotes. Since that point in time, the bacterium has continued to evolveand so have all the other organisms.
If all the eukaryotes have evolved for the same amount of time (from point a to the present), it is not surprising that they are all about equidistant from the bacterium in terms of their cytochrome C sequences. Furthermore, note that the rest of the data in the matrix now make sense. The horse-pigeon difference is small (11 percent) because they split relatively recently (at point f on the time line), while the tunafish differs from
the horse by 18 percent because it split from the horse-pigeon ancestor in the more distant past (at point e). In other words, just what modern science would expect.
Denton's book seems to suggest that even evolutionary scientists must hold humans as the ultimate "goal" of natural selection (in fact, evolutionary theory does not embrace the notion of a predetermined goal or direction). So, perhaps Denton's straw-man evolutionist also thinks that after the bacterium split with the yeast it ceased evolving because its job was done; that the evolution of the yeast halted after the yeast split with the wheat; and so on.
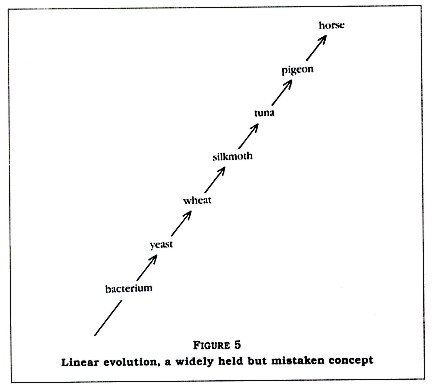
If this were the case, the wheat at point c on the time line would be exactly like the wheat of today. I suspect that Denton's evolutionist also believes that evolution is a linear rather than a branched sequence of events (compare Figure 5 with Figure 4). This would explain why Denton thought he had shocked the scientific community when he pointed out that the difference in the cytochrome C sequence between a bacterium and a wheat is 66 percent and the difference between that bacterium and a tunafish is almost the same. If things had evolved in a linear manner, with bacteria unchanging since point a on the time line, then certainly that bacterium should have a cytochrome C sequence somewhat similar to that of the wheat and much more dissimilar to that of the tunafish.
Denton's confusion in this respect is certainly shared by many other people. Perhaps some of this confusion is the fault of the textbooks that are presently used in our schools (many with little or no discussion of evolution). Perhaps some of it is attributable to our sloppy speech habits: we often hear yeast or bacteria referred to as "primitive" when what we really mean is that "modern" yeast and bacteria have evolved for millions of years to adapt to a changing world but have still retained some degree of their ancestor's simplicity.
Whatever the reason. it seems obvious to me as a teacher that a large portion of the general public has come to accept the notion of linear evolution (for example, that humans evolved from the modern gorilla). We must not let books like Denton's reinforce this notion; we must not let creationists define evolution for the evolutionary scientist.
Acknowledgements
The author would like to thank Dr. Roger Wood, professor of biology at Stockton State College in Pomona, New Jersey, for his helpful comments.